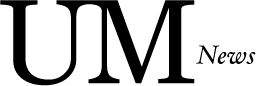From left to right: Julieta Novomisky Nechcoff, Dr. Silvia T. Cardona, Dr. ASM Zisanur Rahman
UM Scientists’ recent discovery opens up possibilities for developing a new antibiotic through AI
Dr. ASM Zisanur Rahman, Julieta Novomisky Nechcoff, and Dr. Silvia T. Cardona have recently published their article, “Rationally designed pooled CRISPRi-seq uncovers an inhibitor of bacterial peptidyl-tRNA hydrolase” in Cell Reports. In this study, the team created a collection of bacterial mutants to help them understand how a new type of antimicrobial molecule, discovered with their artificial intelligence tools, stops bacteria from growing. Their research has now identified a unique combination of a compound and its bacterial target, opening up exciting possibilities for developing a new antibiotic. In this interview, we talk with Cardona, a professor and associate head graduate in the Department of Microbiology and an expert in antibiotic discovery to provide a deeper understanding of her research and recent work.
We have heard concerns about the rise of antibiotic resistance. How does your research address it?
The challenge of antibiotic resistance is a critical one, and new approaches to discovering antibiotics are desperately needed. If you ever wonder how scientists hunt for new antibiotics, you may know some serious detective work is involved to find and understand how new compounds kill infectious bacteria.
Can you expand on how antibiotics kill bacteria?
Yes! An antibiotic kills bacteria by binding to and disrupting a part of their cellular machinery. The target machinery typically performs a process that is essential for survival, so when the antibiotic inhibits this machinery, the cell dies.
How did you apply this concept to your research?
Reducing the abundance of an antibiotic’s target can cause a cell to become hypersensitive to the antibiotic. Dr. A. S. M. Zisanur Rahman, a recent PhD graduate in my lab, applied a clever new technique using CRISPR tools to identify the target of antibiotics by lowering the abundance of different cellular machinery and looking for hypersensitive bacteria. This was quite a lot of work that involved coordination with MSc student Jules Novomisky Nechcoff and Mitacs intern Archit Devarajan.
You mentioned CRISPR, the gene-editing tool. What makes your CRISPR tool different?
We use CRISPR-interference (also known as CRISPRi) instead of regular CRISPR because this technique does not cut DNA, but instead reduces the expression of a gene. If we consider regular CRISPR as a mute button, CRISPRi is like turning down the volume. This allows us to study essential genes without killing the cell we want to analyze.
Why focus on essential genes?
Because targeting essential genes is a good strategy for developing new antibiotics. If you interrupt a critical cellular function, the bacteria can’t survive. Makes sense, right? Hitting them where it hurts.
Can you elaborate on how you applied rational design to build a mutant library?
For sure! Traditional pooled libraries suffer from uneven growth of mutants, leading to the loss of important targets. Zisan built a super-efficient library of CRISPR mutants called CIMPLE, which means: “CRISPRi–mediated pooled library of essential genes”. It’s a fancy way of saying a collection of bacteria, each with a different gene knocked down. By carefully analyzing the growth characteristics of individual CRISPRi mutants, we could predict which mutants would be underrepresented in a pooled system. We used CIMPLE on a completely new growth inhibitor, previously discovered by AI, a mystery drug!
And how did you apply your new tool to antibiotic discovery?
We used CIMPLE on a completely new, uncharacterized growth inhibitor, a mystery drug! We found that the new compound targets a peptidyl-tRNA hydrolase, or Pth, which is an essential bacterial enzyme that helps with protein synthesis. To continue the work on this target, we collaborated with an expert in these types of enzymes: Dr. Yury Polikanov, from the University of Illinois Chicago.
How significant is this research for the field of antibiotic discovery?
The development of CIMPLE provides a more robust and efficient method for identifying novel antibacterial targets, which is crucial given the urgent need for new antibiotics to combat drug resistance. The discovery of Pth as a target for a novel antimicrobial is a direct result of this improved methodology. So, in essence, this paper presents a new, powerful tool for antibiotic discovery, and it identified a new target for antibacterial compounds.
To learn more about Cardona’s research, please visit Cardona Lab’s web page.