Bioinformatics Lab – Q&A with Olivier Tremblay-Savard
“The problem for me always was to choose just one science. They all seemed interesting to me for different reasons,” says Dr. Olivier Tremblay-Savard who ultimately chose to specialize in bioinformatics.
A professor in the department of computer science, Bioinformatics Lab at the University of Manitoba. Dr. Tremblay-Savard’s research interests include comparative genomics and human computing games in genomics. His interest in science goes way back and he credits the terrific science teachers he had in high school/ CEGEP and scientific tv shows for inspiring him. Dr. Tremblay-Savard received a BSc. and a Ph.D. in Bioinformatics from the University of Montreal. He completed a postdoc, in Human Computing and Bioinformatics from McGill University.
Dr. Tremblay-Savard points out that there are many surprises in research, hiding behind every corner, more than we realize. He adds that always keeping an eye out for new opportunities, even after mistakes or failed experiments, may lead to “interesting” and “unexpected” results. When he is not conducting research, Dr. Tremblay-Savard enjoys sports, running, gardening, video games, movies, and cooking.
Kimia Shadkami, a (computer science) graduate student, caught up with Dr. Tremblay-Savard to find out more.
What is your current research focused on? What do you think is significant about it?
My research interests in general, are on all aspects of biological sequence evolution. In other words, how do genomes/sequences change with time, what are the events that cause these changes and how can we infer/identify those evolutionary events. To answer these questions, we need to compare genomes using efficient algorithms because there is a lot of data and the problems are hard. The value in this kind of work comes from both the advancements that we can make in algorithmics and a better understanding of genome evolution that we get from using these algorithms.
With regards to human computing games in genomics, we’re not only interested in developing them to solve a specific problem. We are studying how to best design them, how different features affect player engagement and solution quality, and how to create an experience that is beneficial for the players.
GeSort truly excites me. Tell me about the very first sketch ideas of it. How long have you been working on it? What challenges were you setting out to address?
GeSort is a game about finding the shortest sequence of evolutionary operations to transform one genome into another. The idea for it came from me working on these types of problems during my Ph.D. I found that I often had to look at two genomes and make the moves in my head or on a piece of paper to figure out what was the shortest evolutionary scenario. This was often necessary to check if an algorithm was working properly or not. By doing it so many times I realized it was quite a challenging puzzle!
As soon as I arrived at the University of Manitoba, I started thinking about creating a game for this problem. We have been developing it since 2017 and while we are fine-tuning the game, we are also working on answering questions about player engagement and motivations to help us design the best possible game.
What might the practical outcomes of GeSort be?
The short-term goal with GeSort is to analyze genomes that are evolving too quickly for existing algorithms to be able to produce a good solution in a reasonable amount of time. Ultimately, the long-term goal is to see if we can design better algorithms based on the strategies employed by the players to solve the puzzles.
How do you interpret the data gathered from the game?
That’s a great question. When we study the effects of some game features on the players’ performance and engagement, we can analyze how much time and how many moves were used to solve a specific puzzle, and we also interview the participants to see how much they liked or disliked certain game elements.
When analyzing real genomes, we can compare the solutions produced by players with the solutions produced by existing programs. No existing program is guaranteed to find optimal solutions to the puzzles that we are interested in, because of the complexity of the problem. This means that we can figure out how the human brain’s ability for identifying patterns can help us solve problems that are hard for the computer.
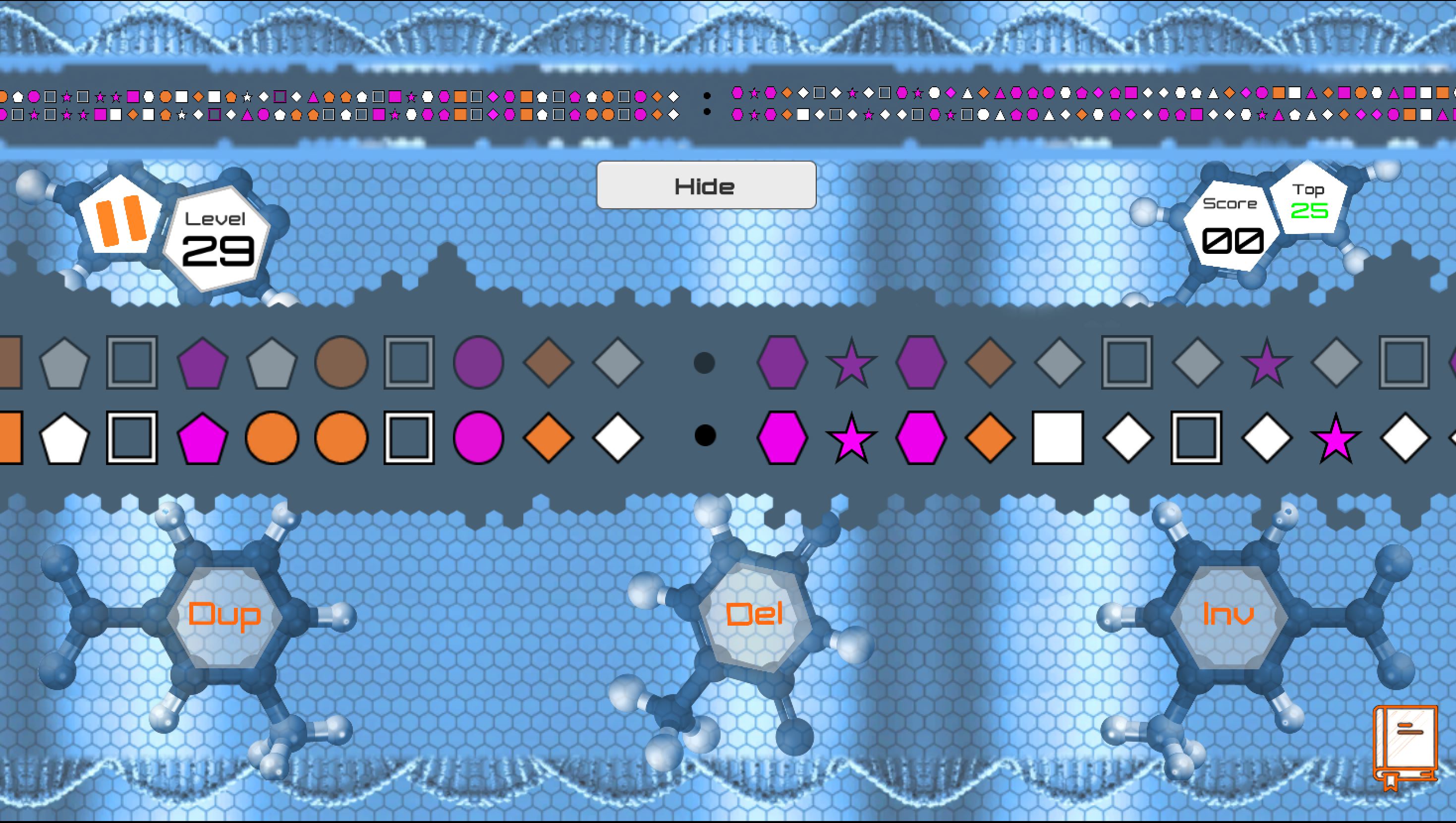
GeSort game.
With humans participating in these gaming experiments are there any concerns you have to address?
That’s an excellent question. I would like to start by mentioning that these kinds of projects involving human participants must always be reviewed and approved by the research ethics board. Before participating in such projects, participants must read and sign (electronically) a consent form that explains what the research is about, the risks and benefits of the project, and the participants’ rights.
Human computing games that aim to solve scientific problems are often called citizen science games. The idea is really to give a chance to everyone to participate in a big science project and be part of the research community. Many studies have shown that citizen scientists are motivated by helping, being part of a big community, and contributing to research.
What is the potential for industry partners?
Recently, citizen science game projects have been integrated into popular commercial video games: Project Discovery in EVE Online and Phylo in Borderlands 3 are good examples of this. These major initiatives have demonstrated the potential for combining the strengths of professional game developers and scientists. The opportunity for big entertainment companies is to leverage the popularity of their video games to advance research and social good. The inclusion of such citizen science mini-games can also help attract more players, or different types of players, to their big commercial games.
I have learned a lot from you and I am sure our readers will do as well. Any advice, kind words, warnings for students or other researchers in this field?
I would say don’t be afraid to follow your passions and interests in research and learn as much as you can in different research areas. Before I chose to do a postdoc on human computing games in genomics, which was going in a different direction than my Ph.D. research, I wasn’t sure if it would be a good idea for my career. I thought: maybe it’s better to stay in the same area and just acquire even more experience in it. The fear back then was to become a “Jack of all trades, master of none.” In the end, it was the best decision I could have made. I think that working on different research topics and areas opens your mind to different ways of approaching problems, and I find that the skills that are acquired in this way can be transferred back to other areas.